PathoSeq
Food safety with high precision – Pathogenomics for the food industry

The objective of the «PathoSeq» project is to prepare Norwegian food industry for the challenges and possibilities of using modern DNA sequencing technologies for surveillance and control of foodborne pathogenic bacteria. The focus is on Listeria monocytogenes, considered the largest food safety challenge in Norwegian food processing industry.
Start
01. Apr 2019
End
31. Mar 2023
Funded by
The Research Council of Norway
Cooperation
The project is a collaboration between Nofima, the Departement of Private Law at the University of Oslo (Norway), the Unit of Food Microbiology at the University of Veterinary Medicine (Vienna, Austria). Industrial partners are Nortura, Grilstad, Norsk Kylling, Orkla Foods Norge, Kjøtt og Fjørfebransjens Landsforbund (KLF), SinkabergHansen, SalMar, Slakteriet, The Norwegian Seafood Federation, Eurofins Genomics, and Aquatiq Sense.
Project Manager(s):
Other Participants:
Even Heir
Merete Rusås Jensen
Solveig Langsrud
Birgitte Moen
Trond Møretrø
Charlotte Kummen
Anette Wold Åsli
Background
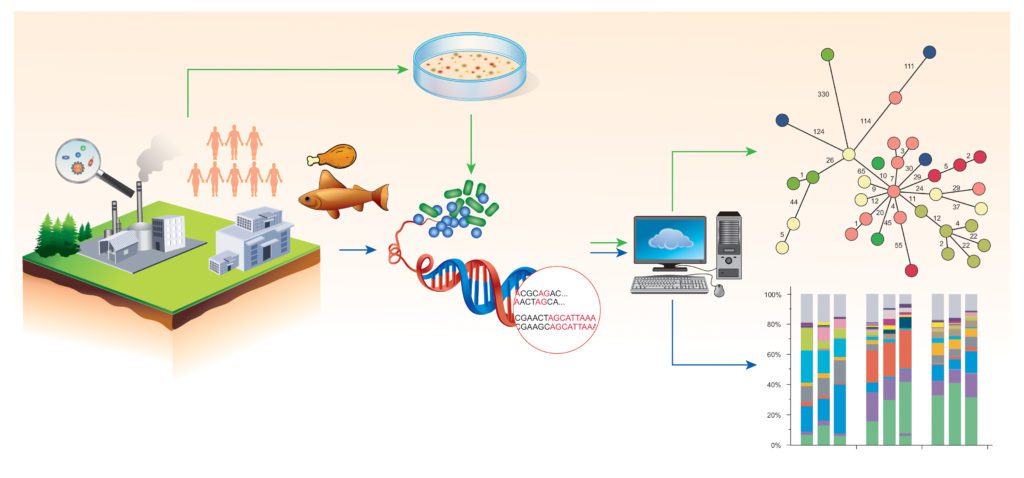
Norwegian food processing industry continuously meets challenges with respect to food safety, and modern food processing requires that sources of contamination with microbial foodborne pathogens can quickly be identified and eliminated from processing facilities. This requires surveillance sampling schemes and methods for discrimination between different bacterial clones.
Whole genome sequencing (WGS) is a method used to determine the complete DNA sequence of an organism’s genome.
WGS has superior sensitivity, specificity and accuracy compared with traditional methods used in surveillance, source tracking, and investigation of foodborne outbreaks. The method is used by public health and food safety authorities during outbreak investigations and for epidemiological surveillance of foodborne pathogens in the food chain, and may enable tracing of potential contaminations back to individual processing plants.
Knowledge and benefits
The project aims to identify opportunities and barriers related to the use of WGS technology in the food industry. The use of WGS technology for preventative contamination controls in the food industry will result in a better understanding of transmission patterns and sources for microbial contamination, giving safer products on the market.
WGS analyses can also provide knowledge about the functional properties of bacteria, such as antibiotic resistance, virulence, and pathogenicity – knowledge that may be used to improve food safety management and risk assessment plans.
Project results
The project has sequenced nearly 800 environmental isolates of Listeria monocytogenes from 15 different Norwegian food processing plants, collected over a period of 30 years. This has resulted in increased knowledge of the origin, diversity and distribution of L. monocytogenes in Norwegian food chains.
Mapping of genetic relationships between the individual isolates showed that over half of the isolates were very closely related to bacterial isolates collected from different factories. The genetic similarity between isolates from different factories were at the same level as that often seen among isolates linked to the same listeriosis outbreak. Consequently, a broad overview over the L. monocytogenes clones circulating in a food chain is important background knowledge needed for interpretation of WGS results, for example, during investigations of foodborne outbreaks.
Examination of genetic determinants showed that the dominant L. monocytogenes variants established as persistent house strains in more than one factory were adapted to the production environment, and more frequently carried genes that provide resistance to various types of stress, including cleaning and disinfection.
Investigation into the use of WGS data to identify specific genes associated with disease severity was performed to examine the possibility of using this information as part of risk-based food safety management in the industry. The results showed that this approach currently has limitations, as the virulence gene profiles did not always correlate with experimental data for invasion efficiency determined using cell assays.
Furthermore, the potential of metagenomic sequencing based methods for faster and more sensitive analysis results has been examined. The results showed that it was possible to detect L. monocytogenes after four hours of enrichment using Nanopore sequencing technology. This approach may also be used to characterize the background microbiota in environmental samples. In addition, sequencing can be used to investigate whether there is more than one variant of L. monocytogenes present in a sample.
Practical analysis strategies and regulatory aspects
The project has been involved in development of tools for visualization and handling of WGS results in the food industry. We will prepare practical guidelines for how food business operators may use and integrate WGS data into their food safety management systems.
The project employs a transdisciplinary and multi-actor approach to produce and translate science into practical recommendations for the food industry. This will include clarification of legislative and other regulatory aspects relevant for implementation of WGS methodology for regulation and control of food safety, at both domestic and international levels. This work is led by the University of Oslo.
Publications
Topics
Pathogens